About
I am a fourth-year Biology Ph.D. candidate at Texas A&M University. I use bioinformatics, population genetics, and molecular genetic tools to probe the role of genome structural variation in adaptive evolution.
- Email: asamano@tamu.edu
- Advisor:Dr. Mahul Chakraborty
Research Interests

Adaptive Evolution
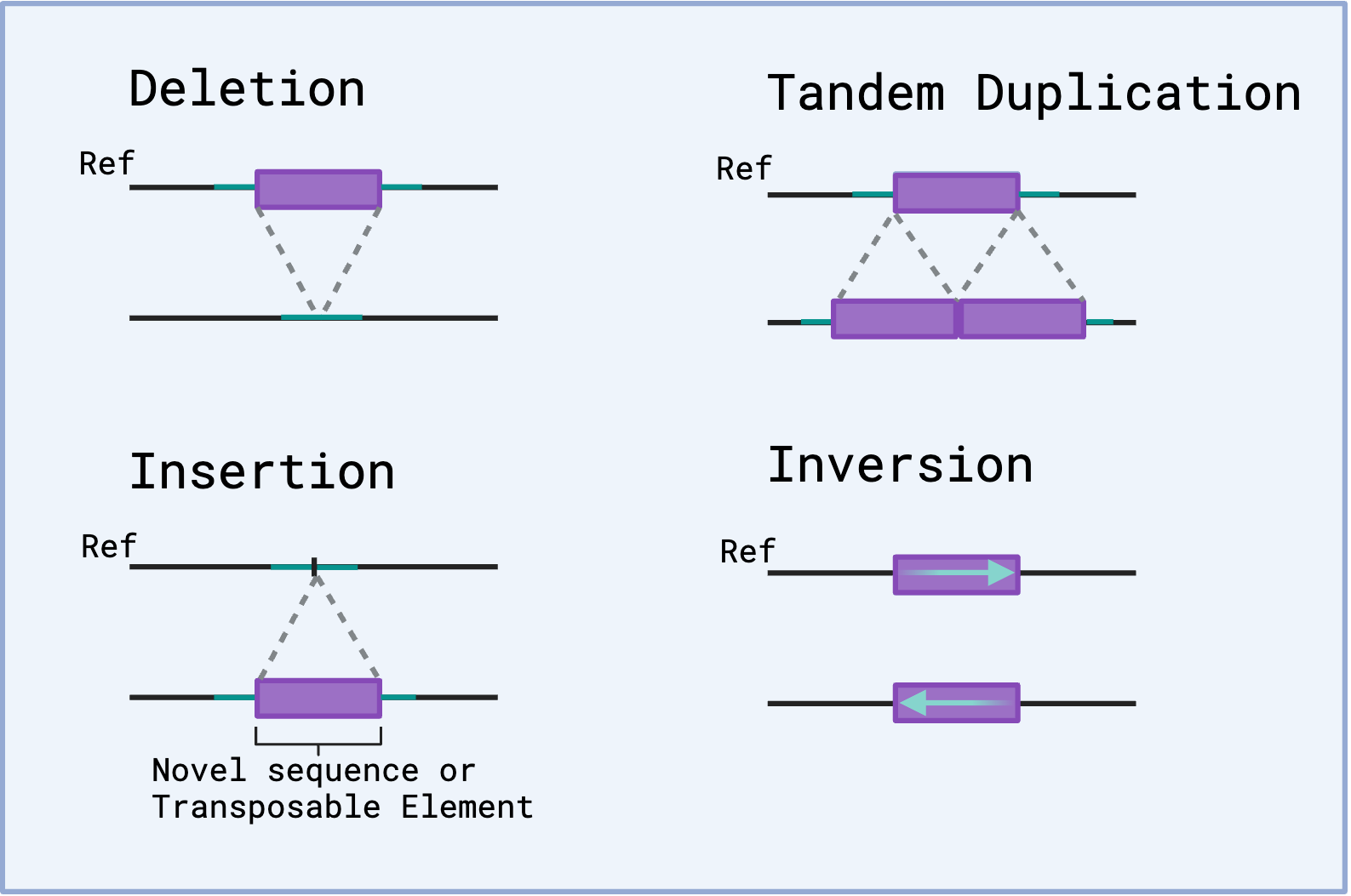
Genome Structural Variation
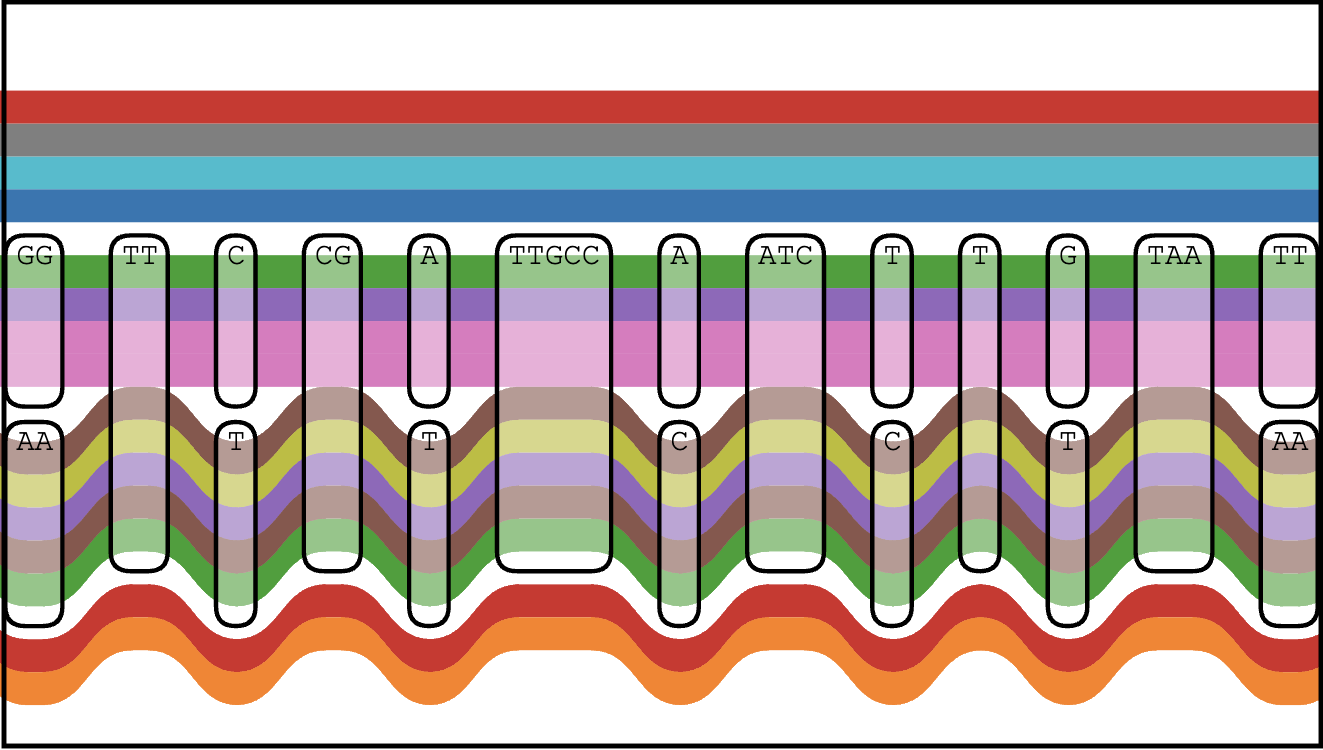
Pangenomics
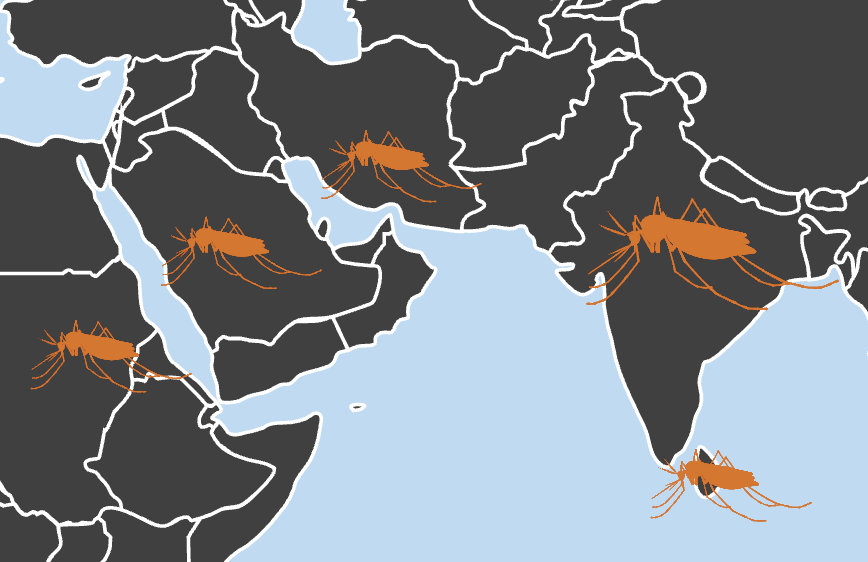
Invasive Species
Publications
Alejandra Samano, Matthew Musat, Mihir Junaghare, Asad Ahmad, Mehlum Ali, Sebastian Alves, Sreeram Pasupuleti, Jelisha Perera, Omar Saada, Brady Sabido, Trevor Smith, Sophie Walz, Mahul Chakraborty.
Structural variants are enriched in deleterious visible phenotypes in Drosophila
Under revision.
Alejandra Samano, Naveen Kumar, Yi Liao, Farah Ishtiaq, Mahul Chakraborty.
Genome structural variants shape adaptive success of an invasive urban malaria vector Anopheles stephensi.
In: Molecular Biology and Evolution, Volume 42, Issue 6, June 2025.
Teaching
BIOL450 Genomics, Spring 2024, 2025, 2026, Texas A&M University: Instructor for lab section of upper-level biology course. Students learn how to perform genome-assembly, comparative genomics, and structural variant annotation.
BIOL112 Introductory Biology II, Spring 2023, Texas A&M University: Teaching assistant for general biology laboratory sections covering plant and animal biology.
BIOL111 Introductory Biology I, Fall 2022, Texas A&M University: Teaching assistant for general biology laboratory sections covering basic principles in cell biology.
Education and Professional Experience
Education
Texas A&M University - College Station
Biology Ph.D.
August 2022 - present
Graduate student in the lab of Dr. Mahul Chakraborty. My current project involves identifying SVs in geographically diverse Drosophila melanogaster genomes with the goal of understanding how repetitive genomic elements contribute to adaptive evolution.
University of North Carolina at Chapel Hill
Bachelor of Science in Biology
Minor in Chemistry and Computer Science
August 2016 - May 2020
Undergraduate Researcher with Dr. Christopher Willett's lab studying evolutionary genetics of speciation. Worked with bioinformatic programs to analyze the genome of a copepod species, Tigriopus californicus.t>
Professional Experience
Genomics Lab Supervisor
Mako Medical Laboratories, Henderson, North Carolina
January - July 2022
- Supervisor of clinical laboratory testing for SARS-CoV-2.
Molecular Lab Scientist I
Mako Medical Laboratories, Henderson, North Carolina
July 2020 - January 2022
- Trained and certified in all major steps of processing COVID-19 samples: RNA extraction, qPCR, and analysis of gene expression data to identify the presence of SARS-Cov-2 and variants.
Contact
Check out my lab's website here or reach out to me for details if you are interested in participating in research! I enjoy mentoring undergraduates who want to get research experience in genetics, evolution, and bioinformatics.
Email:
asamano@tamu.edu
